The previous article in this series is Using Genome Mate (Add confirmed segments to chromosome map).
As explained in my earlier post, Using Genome Mate (Identify triangulated groups and confirm segments), we now have an understanding of the basic principals of triangulating DNA segments, and know how to manually go through our match data and identify triangulated groups. In this post, I will show how to use the GEDmatch Triangulation tool to automatically identify the triangulated segments you share with your 400 closest matches at GEDmatch and then easily import the triangulated segment results from GEDmatch into Genome Mate.
Click here if you’d like to open the GEDmatch website in a new window/tab to make it easier to follow along with these steps.
In order to use the Gedmatch Triangulation tool, you need to be a subscriber to the GEDmatch Tier 1 service. The cost for the Tier 1 service is $10 per month, and you can subscribe in increments as little as a month at a time. Tier 1 service is well worth the nominal cost, even if you use it for nothing more than the Triangulation tool. In addition to the Triangulation tool, however, Tier 1 service will also provide you access to the Matching Segment Search tool, the Relationship Tree projection tool, and the Lazurus tool, and the use of these three tools will be covered in future posts. The instructions for subscribing to the Tier 1 service are located at the bottom of the GEDmatch main page (http://www.gedmatch.com), after you’ve signed in.
After subscribing to the Tier 1 service, and signing back in to the GEDmatch website, you will see an additional blue box at the lower right of the GEDmatch main page showing the Tier 1 Utilities.
1) In the Tier 1 Utilities box, click on the Triangulation tool link.
2) Clicking on the Triangulation tool link will bring up the GEDmatch Segment Triangulation page. Enter your kit number in the first box, select No Limit in the Upper Segment Threshold Limit dropdown, click the “Show results sorted by chromosome, segment start position” radio button, and then click the Triangulate button at the bottom.
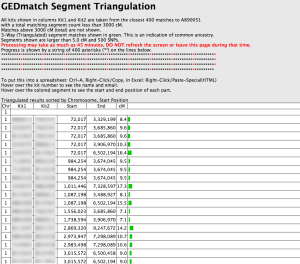
3) After clicking the Triangulate button, you will get a new page which says “Progress is shown by a string of 400 asterisks (‘*’) on the lines below:” on the last line.
Asterisks will begin slowly appearing in black, with every tenth asterisk appearing in red. This is the indication that the Triangulation tool is processing your results. The processing can take up to forty-five minutes to completely finish. It is very important to let this processing finish, and not interrupt it by clicking the stop or refresh buttons on your browser, closing your browser, or navigating to another page in your browser. If you do happen to interrupt this processing, open a new browser window/tab or restart your Internet browser, log back in to the GEDmatch page (http://www.gedmatch.com), and repeat steps 1 through 3 above.
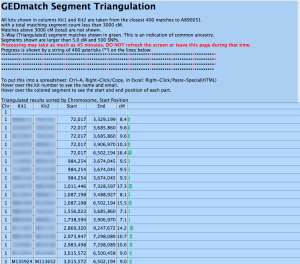
Once the Triangulation tool has completely finished processing, you will receive a results page that resembles the following screenshot.
4) While viewing your results screen, select everything on the page (Windows users hold down the Control key and press the A key on your keyboard, Mac users hold down the Command ⌘ key and press the A key on your keyboard). Copy the data (Windows users hold down the Control key and press the C key on your keyboard, Mac users hold down the Command ⌘ key and press the C key on your keyboard), and then switch to your Genome Mate software.
5) In Genome Mate, after making sure you have first backed up your database, click on the Import Data link in the menu bar near the top of the main Genome Mate window.
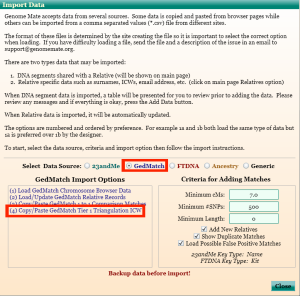
6) Clicking on the Import Data link will bring up the Import Data window. Select the radio button next to GedMatch in the Select Data Source section, and then click on the (4) Copy/Paste GedMatch Tier 1 Triangulation ICW link in the GedMatch Import Options section.
7) Clicking on the (4) Copy/Paste GedMatch Tier 1 Triangulation ICW link in the GedMatch Import Options section will bring up another Import Data window.
8) Click once inside the rectangular box, and then paste the data you previously copied from GEDmatch (Windows users hold down the Control key and press the V key on your keyboard, Mac users hold down the Command key and press the V key on your keyboard). After pasting your data into the box, click on the Format button. [Note: Although the instructions warn that only the Chrome and Firefox browsers are supported, I have found that as of Genome Mate version 1.2.19, the Safari browser also works in most cases.]
9) Clicking on the Format button will bring up the Import Summary window. This acts as a confirmation that the data was imported, as long as ICW records processed is not 0 (zero). If ICW records processed is zero, this is letting you know that you have previously imported all these ICW records, or is an indication that something went wrong with your import.
If you have not imported these ICW records before, and something seems to have went wrong with your import, you should click the Close button and repeat steps 4 through 8 above. If you have repeated steps 4 through 8 above, and are still getting a zero for ICW records processed, please let me know by adding a comment to this blog post, or by sending me an email, so that I can help you determine what is causing the problem you are experiencing.
After having imported your ICW records, click the Close button.
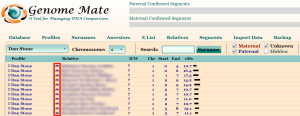
After clicking the Close button, you will be taken back to the main Genome Mate window. It is a good idea to get in the habit of backing up your Genome Mate database after each import you do. In front of most, if not all, of your matching kits from GEDmatch, you will notice a small symbol that looks like four people standing together. This is the indication that ICW (in common with) data exists for that match.
Moving your mouse pointer over one of these symbols will highlight all the other matches of yours who also match the person whose symbol your mouse pointer is currently over.
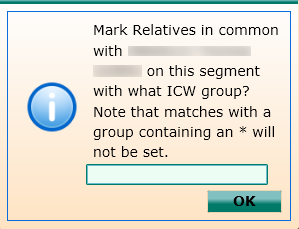
Clicking on the symbol will then bring up a box that allows you to set the ICW Group for all of these highlighted segments (for an explanation of the principles of setting ICW Groups, please see my post Using Genome Mate (Add confirmed segments to chromosome map).
If you know whether the highlighted group matches on the paternal or maternal side of your family tree, it is best to correspondingly mark the highlighted segments as either P or M. If you don’t yet know whether the highlighted group matches on the paternal or maternal side of your family tree, however, you can still make use of this feature.
Instead of marking the ICW Group as either P or M, instead mark all the highlighted segments with a designation you’ve decided upon that represents the group has been triangulated but is still undetermined as to which side of your tree the group matches you on. For example, I use the letters A and B. You could also use the numbers 1 and 2. Anything will work for this purpose, other than using the letters P and M, or an asterisk (*).
Here, I have marked all the highlighted segments as A.
Any segments that line up vertically with the segments I’ve just marked as ICW Group A, but aren’t part of ICW Group A, I mark with ICW Group B, since I know it is extremely likely they will match on the opposite side of my family tree from the side ICW Group A matches me on.
This lets me know, as I look through my chromosomes in Genome Mate, that I’ve identified the triangulation group, but still need to work on finding out which side of my family tree the group matches me on.
Once I am able to determine the correct side (paternal or maternal) a group matches me on, however, I can mark all of the segments in that group as either ICW Group P or ICW Group M, whichever the case may be. I then mark the other group I previously designated as the ICW Group of the opposite side of my family tree.
For example, let’s say that I discover a first cousin on my mom’s side of my family tree shares the same segment of chromosome 1 with me as with one of my matches from ICW Group B. I can then mark ICW Group B as M, and mark ICW Group A as P.
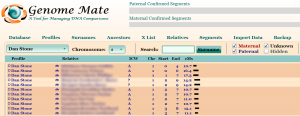
If you are fortunate enough to have been able to test a parent and/or grandparent, the Triangulation tool simplifies things even further. In the example shown below, I use the symbol in front of my mother’s test on the first line to highlight and mark the ICW Group as M of all the matches shared between me and my mother.
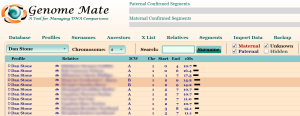
In the next example shown below, I use the symbol in front of my paternal grandmother’s test on the third line to highlight and mark the ICW Group as PGM of all the matches I share with my grandmother. [Note: After marking all of the segments shared with my paternal grandmother as PGM, I go back and set the ICW Group of my father’s test on the second line to correctly be just P, rather than PGM, since the matches I share with my father come from both sides of his ancestral family tree.]
I hope this information is helpful. Using the GEDmatch Triangulation tool, in conjunction with Genome Mate, should greatly speed up your identification of triangulated groups of segments, allowing you to more efficiently begin working with the matches in these groups to try to identify the common shared ancestor(s) for each group.
As with all my posts, if you experience any problems while following these steps, notice that a screen has changed dramatically from what I’ve shown in the screenshots, or have an easier/better way to accomplish what I’ve shown, please let me know by commenting on this thread or sending me an email. Thank you.
This article last updated 10 Jan 2015















Pingback: Steps to DNA Matching and Chromosome Mapping with GEDmatch and Genome Mate | Adventures In Genealogy Research: No Stone Unturned/The Wright Stuff
Pingback: Using Genome Mate (Add confirmed segments to chromosome map) | Adventures In Genealogy Research: No Stone Unturned/The Wright Stuff
I have encountered 2 problems with setting up Genome Mate. I am using it for the 1st time and I first attempted to upload the Family Finder Match and ICW files from FTDNA. The ICW file would not upload as all files rejected due to format.
I then attempted to upload the Gedmatch Tier 1 Triangulation file (copy/paste) and as you mentioned in your article I received a resulting 0 files processed. I “deleted everything” started anew and again tried to import the Triangulation file with the same result. I repeated this 4 times without success.
Can you help me?
Appreciate your articles.
David
Hi David,
Thanks for the information you’ve supplied regarding the problems you are encountering. I want to ask a couple questions in order to see if we can determine specifically what may be causing these problems:
When you attempted to import your FTDNA data, did you use the DNAGedcom site to download the files?
When you say that you “deleted everything” does this mean that you have started a new database to test the imports to see if they work? If not, I would suggest making a backup of your current database, create a new database, do a new download of your FTDNA data through the DNAGedcom site, and then try importing the three files into Genome Mate in the order of Family Finder file first, Chromosome Browser file second, and ICW file third. After trying to import these three files, regardless of whether you are successful or not, try to import the GEDmatch Tier 1 Triangulation data again.
Let me know the results of trying this, and if you get any error messages. If you are still having problems I can provide some further things to try based on the error messages you get.
Dan
Hi Dan. For some reason my import results in zeros.
I took a screen shout, but apparently I can’t attach it.
I just tried another one of my relatives with the same zero results. I notice that in both cases, the final red asterisk does not appear; however, when I examine the data, it looks complete. I used option (4) as you instruct. Here are the results after I click the format button:
Matches added: 0
Relative records added: 0
Total import count: 2,295
Rejected for invalid format: 1,549
ICW records processed: 746
I am accessing Genome for a first as well and am getting”
0 Matches
0 Relatives
3117 Total Import
3112 Rejected for invalid format
5 ICW Processed
However I get nothing when I click close
Hi Joe and Alicia,
It sounds like both of you are doing everything correctly. In regards to the final red asterisk not appearing, this is normal, as it jumps immediately to displaying the results at that point rather than displaying the last asterisk before moving on to displaying the results. Since both of you are not getting zeros for ICW processed, something does seem to be getting imported. Alicia’s result of five, however, seems to be extremely small, so I’m curious to know if the GEDmatch triangulation results page looked like it had just a very small number of green segments that are over 7 cM (or whatever threshold you set).
Thanks,
Dan
I don’t know if this will help but I was having a similar problem while signed onto gedmatch through Internet Explorer. I loaded the Chrome Browser and signed into gedmatch on there and everything worked.
Thank you, Carolyn. This is very helpful to know, and I appreciate you sharing what ended up working for you for the benefit of anyone else who may run into this same issue.
Thanks for the Genome Mate articles! I’ve been using it for over a month but learned several things from your articles I hadn’t noticed or figured out before.
One thing to remember before marking all your non-A segments as B: Tier1 Triangulation only imports your top 400 matches out of 1500. That’s quite a few segments that don’t get checked at all. I have a child and one-parent combo, and I almost made the mistake of — after marking the child’s segments that match the mom as M — marking the remaining segments as P. That would have made a mess with lots of mistaken assumptions! They have to be checked manually first. (And then of course I did mark all the remaining ones that didn’t manually match the mother, as P). I wish there was a way to check all 1500 automatically!
Hi Walker,
Thanks for your comments, and good advice. Yes, it would be fantastic to have a tool that automatically triangulates all 1500 of one’s matches. They have already upped the number of matches triangulated by the tool from 300 to 400, so at some point in the future perhaps this will extend to one’s entire match list. Here’s hoping, at least.
Best regards,
Dan
Thanks so much for your helpful instructions about the use of Genome Mate. I was eager to try it, but I can’t get the table to populate with data.
I use an IMac with Version OS 10.7.5. Although I normally use Chrome, I installed Silverlight with Safari as I had read Chrome was no longer allowing the Silverlight plug-in. I had to enable a security work- around in Safari in order to install Silverlight, but after doing so, it installed just fine on my machine.
I downloaded Genome Mate in Safari and went through the entire process of creating a profile. I used Chrome to create the GedMatch Tier 1 Segment Triangulation results. I opened Genome Mate to the main page, hit the Import Data button, the Import Data window opened, and I pasted in the GedMatch Triangulation results copied from my Chrome browser. I hit the Format button and the Import Summary page showed that 1,739 records had been processed.
I closed the Import Summary page and the Import Data page but there appeared no data in the main window of Genome Mate. Has anyone else had this occur? Is there something I failed to do along the way. The data appears to have been transferred to Genome Mate, but it won’t populate the table.
Thanks in advance for any advice.
Hi C Hartsough,
Sorry to hear of the trouble you seem to be having. I can think of a few things to try:
1) If you have more than one profile setup in your Genome Mate database, try switching to another profile, and then switch back to the profile into which you imported the GEDmatch Triangulation data. For some unknown reason, I have noticed that the segments don’t always appear in Genome Mate until I have switched profiles. If you don’t have another profile in your Genome Mate database, create a temporary profile and try the steps I’ve just outlined.
2) Click on the Relatives menu item, and see if there are any GEDmatch keys listed. If so, this is an indication that the GEDmatch data is being imported, and that the problem is some kind of issue with the displaying of the segment data.
3) If neither of the above seems to work, try doing a GEDmatch one to one import.
If none of the above work, please let me know what steps you tried and what the result was, so we can troubleshoot further.
Hope this helps.
Best regards,
Dan
1) Click on “Profiles,” then next to “Select Profile,” choose your profile in the drop-down list. You will see “Gedmatch Kit #” — Make sure you have entered your Gedmatch number properly.
2) Next, when importing data, I would recommend using option (1b) Tier1 Matching Segment Search data option first. This is the only option (other than 1-to-1 import) that imports SNPs; any other option will just put “700” as the value, and even if you import later using 1-to-1 or Matching Segment Search, that 700 will not be updated to the real value.
3) Next, use option (2) Load/Update Gedmatch Relative Records
4) Now you can use option (4) Triangulation data import. NOTE that during this import, GemomeMate warns that “Only data for existing relatives will be added. This should be the last step in adding data.” I will add that you can do option (3) for 1-to-1 import anytime. But it is recommended you do the others in the numerical order they are presented in GenomeMate.
Hope this helps!
Dan and Walker,
Success!
Thanks very much for your recommendations about getting segments from an import with Gedmatch to appear in Genome Mate. After trying several, the only strategy that really seemed to work was importing the data from Gedmatch in strict order. In other words when I imported Tier1 Matching Segment Search data first (option 1b) and then imported Tier1 Segment Triangulation data (option 4), the segments appeared in the Genome Mate table. The other way around, no.
Sincerely,
C. Hartsough
I’m so glad to hear you were able to get this resolved. Thanks very much to Walker for assisting with this issue!
So, I’ve loaded all my matches from gedmatch and once I assigned my mother as “M” she disappears from the list, but shows up in my son’s profile (segments overlapping Garrett on GedMatch by at least 5). Would all in this box be born from the triangulation data I imported? Also, am I setting the MRCA to my mother by assigning an “M” to each chromosome she appears on and if that is the case why wouldn’t the triangulation feature add the ICW?
Bill
Hi Bill,
I suspect the reason why your mother disappeared from the list as soon as you assigned her the M designation is because the Maternal checkbox may not be checked, which caused any segments beginning with the M designation to be hidden from view in the main Genome Mate window. I’m not positive I’m sure which box you are referring to in your question, but I believe you are meaning the Relative Record screen. If so, the information all comes from the various imports to Genome Mate, and not just from the triangulation data that is imported. For example, I have profiles for myself and both my parents, so it will show me someone who matches both me and one of my parents on the same segment as a triangulated match, without me ever having imported the triangulation data. Likewise, while Genome Mate does many helpful things, it will not set the ICW since it has no way of knowing the specific relationship between two profile persons.
I hope this helps. However, if I’ve not understood your questions correctly, please let me know a bit more information about your specific situation, and the screen(s) you are looking at in Genome Mate, so that I can properly answer your questions for you.
Best regards,
Dan